Visualizing nanoscale structures in real time
Open-source software enables researchers to see materials in 3D while they’re still on the electron microscope.

Open-source software enables researchers to see materials in 3D while they’re still on the electron microscope.
U-M EXPERTS:
Computer chip designers, materials scientists, biologists and other scientists now have an unprecedented level of access to the world of nanoscale materials thanks to 3D visualization software that connects directly to an electron microscope, enabling researchers to see and manipulate 3D visualizations of nanomaterials in real time.
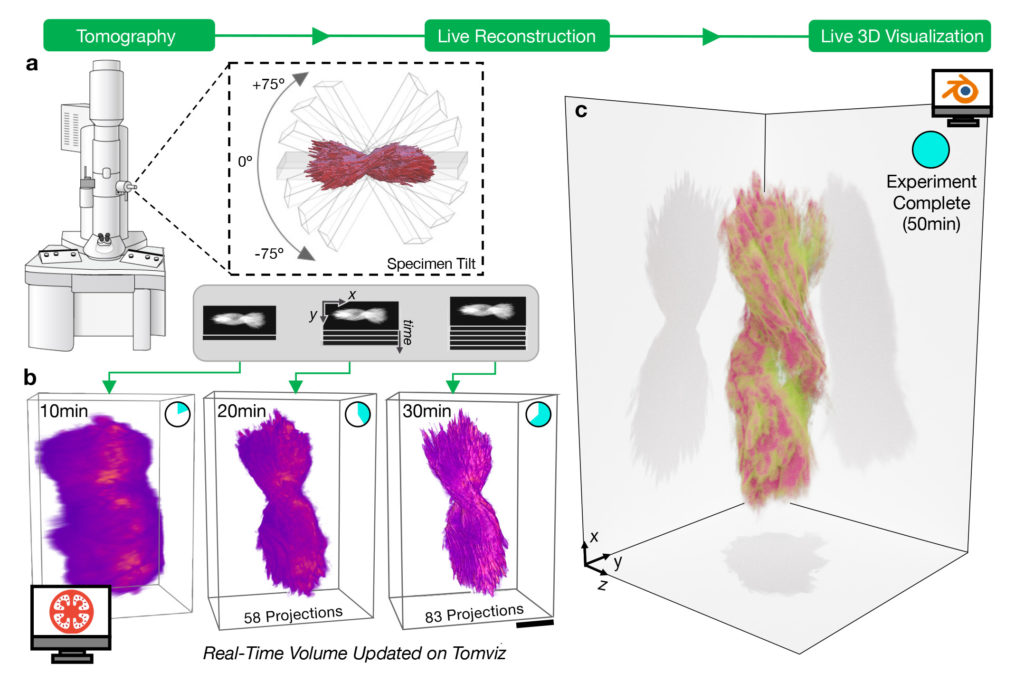
Developed by a University of Michigan-led team of engineers and software developers, the capabilities are included in a new beta version of tomviz, an open-source 3D data visualization tool that’s already used by tens of thousands of researchers. The new version reinvents the visualization process, making it possible to go from microscope samples to 3D visualizations in minutes instead of days.
In addition to generating results more quickly, the new capabilities enable researchers to see and manipulate 3D visualizations during an ongoing experiment. That could dramatically speed research in fields like microprocessors, electric vehicle batteries, lightweight materials and many others.
“It has been a longstanding dream of the semiconductor industry, for example, to be able to do tomography in a day, and here we’ve cut it to less than an hour,” said Robert Hovden, an assistant professor of materials science and engineering at U-M and corresponding author on the paper, published in Nature Communications. “You can start interpreting and doing science before you’re even done with an experiment.”
Hovden explains that the new software pulls data directly from an electron microscope as it’s created and displays results immediately, a fundamental change from previous versions of tomviz. In the past, researchers gathered data from the electron microscope, which takes hundreds of two-dimensional projection images of a nanomaterial from several different angles. Next, they took the projections back to the lab to interpret and prepare them before feeding them to tomviz, which would take several hours to generate a 3D visualization of an object. The entire process took days to a week, and a problem with one step of the process often meant starting over.
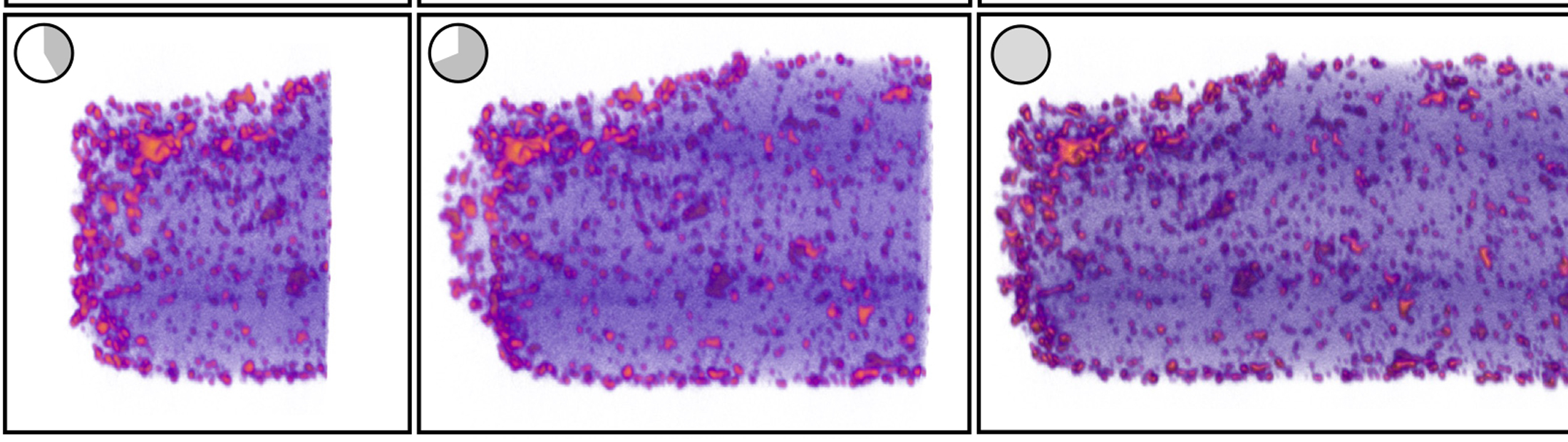
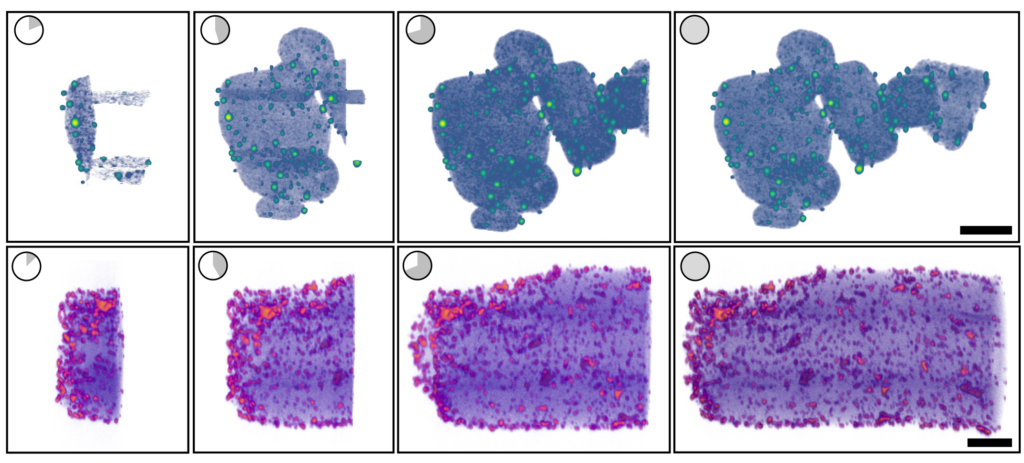
The new version of tomviz does all the interpretation and processing on the spot. Researchers get a shadowy but useful 3D render within a few minutes, which gradually improves into a detailed visualization.
“When you’re working in an invisible world like nanomaterials, you never really know what you’re going to find until you start seeing it,” Hovden said. “So the ability to begin interpreting and making adjustments while you’re still on the microscope makes a huge difference in the research process.”
The sheer speed of the new process could also be useful in industry—semiconductor chip makers, for example, could use tomography to run tests on new chip designs, looking for failures in three-dimensional nanoscale circuitry far too small to see. In the past, the tomography process was too slow to run the hundreds of tests required in a commercial facility, but Hovden believes tomviz could change that.
Hovden emphasizes that tomviz can be run on a standard consumer-grade laptop. It can connect to newer or older models of electron microscopes. And because it’s open-source, the software itself is accessible to everyone.
“Open-source software is a great tool for empowering science globally. We made the connection between tomviz and the microscope agnostic to the microscope manufacturer,” Hovden said. “And because the software only looks at the data from the microscope, it doesn’t care whether that microscope is the latest model at U-M or a twenty-year-old machine.”
To develop the new capabilities, the U-M team drew on its longstanding partnership with software developer Kitware and also brought on a team of scientists who work at the intersection of data science, materials science and microscopy.
At the start of the process, Hovden worked with Marcus Hanwell of Kitware and Brookhaven National Laboratory to hone the idea of a version of tomviz that would enable real-time visualization and experimentation. Next, Hovden and Kitware’s developers collaborated with U-M materials science and engineering graduate researcher Jonathan Schwartz, microscopy researcher Yi Jiang and machine learning and materials science expert Huihuo Zheng, both of Argonne National Laboratory, to build algorithms that could quickly and accurately turn electron microscopy images into 3D visualizations.
Once the algorithms were complete, Cornell professor of applied and engineering physics David Muller and Peter Ericus, a staff scientist at the Berkeley Lab’s Molecular Foundry, worked with Hovden to design a user interface that would support the new capabilities.
Finally, Hovden teamed up with materials science and engineering professor Nicholas Kotov, undergraduate data scientist Jacob Pietryga, biointerfaces research fellow Anastasiia Visheratina and chemical engineering research fellow Prashant Kumar, all at U-M, to synthesize a nanoparticle that could be used for real-world testing of the new capabilities, to both ensure their accuracy and show off their capabilities. They settled on a nanoparticle shaped like a helix, about 100 nanometers wide and 500 nanometers long. The new version of tomviz worked as planned; within minutes, it generated an image that was shadowy but detailed enough for the researchers to make out key details like the way the nanoparticle twists, known as chirality. About 30 minutes later, the shadows resolved into a detailed, three-dimensional visualization.
The source code for the new beta version of tomviz is freely available for download at GitHub. Hovden believes it will open new possibilities to fields beyond materials-related research; fields like biology are also poised to benefit from access to real-time electron tomography. He also hopes the project’s “software as science” approach will spur new innovation across the fields of science and software development.
“We really have an interdisciplinary approach to research at the intersections of computer science, material science, physics, chemistry,” Hovden said. “It’s one thing to create really cool algorithms that only you and your graduate students know how to use. It’s another thing if you can enable labs across the world to do these state-of-the-art things.”
Kitware collaborators on the project were Chris Harris, Brainna Major, Patrick Avery, Utkarsh Ayachit, Berk Geveci, Alessandro Genova and Hanwell. Kotov is also the Irving Langmuir Distinguished University Professor of Chemical Sciences and Engineering, Joseph B. and Florence V. Cejka Professor of Engineering, and a professor of chemical engineering and macromolecular science and engineering.
“I’m excited for all the new science discoveries and 3D visualizations that will come out of the material science and microscopy community with our new real-time tomography framework,” Schwartz said.