Toward protein nanomachines: just add charge
Added electrical charges can harness a protein’s shape and chemical properties to build interesting structures.

Added electrical charges can harness a protein’s shape and chemical properties to build interesting structures.

By putting electrical charges on proteins, it may be possible to make them assemble according to a design, researchers at the University of Texas at Austin and the University of Michigan have shown. The end goal of work in this vein is protein nanomachines, which could deliver drugs directly to tumors or turn sunlight into fuel.
“We think we can use these structures kind of like Legos to build bigger things,” said David Taylor, assistant professor of molecular biosciences at UT Austin and co-corresponding author on a new paper published in the journal Nature Chemistry. “We also know some of the rules for modifying the basic recipe to make different types of building blocks.”
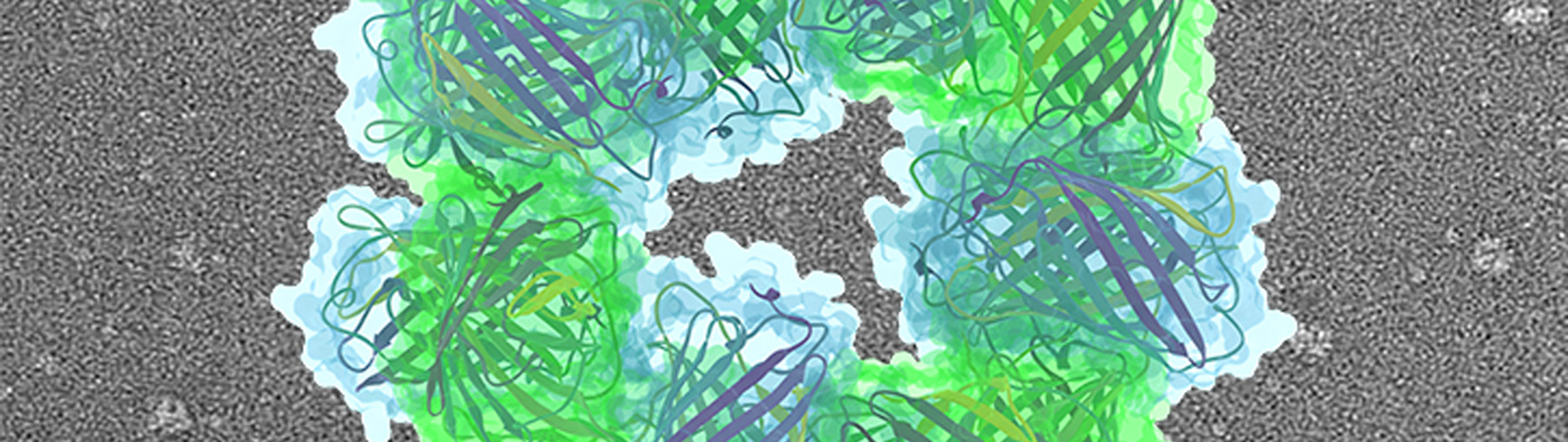
As a proof of concept, the team built tiny structures that resemble two doughnuts stacked on top of each other. They coaxed naturally occurring proteins to form these structures by applying electrical charges to specific spots.
“It’s like a first baby step toward engineering very complex structures of proteins by design,” said Sharon Glotzer, the Anthony C. Lembke Department Chair of Chemical Engineering at U-M. Glotzer and Jens Glaser, an assistant research scientist in chemical engineering, used the data gathered by the UT Austin team to verify their computational model, which may eventually guide the design of more ambitious protein structures.
Earlier methods of creating synthetic protein structures are complicated and take a long time. Researchers made new proteins from scratch or painstakingly affixed molecules onto the proteins to force them to assemble in a particular way. By contrast, the new method is flexible and relies on self-assembly, mimicking the way that proteins in living organisms work as they make the molecular machines that carry out the different functions of life.
“It’s like a first baby step toward engineering very complex structures of proteins by design.”
Sharon Glotzer, the Anthony C. Lembke Department Chair of Chemical Engineering
It was a bit of a gamble because the method relies on the properties of the protein itself to direct self-assembly—simply adding positive and negative charges to objects could potentially result in a shapeless mass. But the protein’s own chemical and shape complexity meant that certain configurations were more favorable than others, resulting in interesting structures.
“Our approach takes a protein that doesn’t normally assemble, and gives it many potential sites where it might be able to, allowing it to ‘choose’ which fits the rest of its geometry and chemistry best,” said Anna Simon, a postdoctoral researcher in molecular biosciences at UT Austin and co-first author of the paper. “This is important because it gives us a way to semi-direct proteins to organize into larger structures without having to understand beforehand exactly how they will fit together.”
To demonstrate their concept, the researchers started with green fluorescent protein, a standard protein used as a glowing tag in biological experiments. They put positive charges on one version and negative charges on the other by adding acidic or basic amino acids to the protein surfaces. The acidic amino acids create positive charges because they want another electron, whereas the basic amino acids are like negative charges, willing to give up an electron.
Mixed together, the proteins self-assembled into tiny structures, yielding many copies of the “stack of two donuts”. A ring of eight proteins—alternating positively and negatively charged proteins—bonded with another ring of eight proteins to create a reliable 16-protein structure.
Because this method allows structures to be built from naturally occurring proteins, the researchers say it offers science a new tool that’s scalable, affordable and sustainable.
“It’s like how people use 3D printers to make things out of materials they wouldn’t have used in the past,” Taylor said. “This new method gives us another option for materials. These materials are easily obtainable, inexpensive and not harmful to the environment.”
Rather than trying to model every facet of the protein’s structure, the U-M team represented every protein as a detailed, three-dimensional surface studded with electrical charges. Vyas Ramasubramani, a graduate student in chemical engineering at U-M, performed simulations of the model on the supercomputer “Stampede2”, which is also located at UT Austin.
“Our shape-based model is so simple that we could study the entire protein complex—not just a single protein,” said Glaser. “Yet, even though our minimal representation may seem less sophisticated than some more detailed ones which would take years to simulate, our model could still tell the difference between unstable structures that wouldn’t show up in an experiment and the stable structures observed by our colleagues at UT Austin.”
The original concept for this new “supercharged” method of protein synthesis was developed by Andy Ellington, associate director of UT Austin’s Center for Systems and Synthetic Biology, also a professor of molecular biosciences and the co-corresponding author on the study. Yi Zhou, a postdoctoral researcher in UT Austin’s Department of Molecular Biosciences is co-first author of the paper.
David Taylor is a CPRIT Scholar supported by the Cancer Prevention and Research Institute of Texas. Sharon Glotzer is also John Werner Cahn Distinguished University Professor of Engineering, Stuart W. Churchill Collegiate Professor of Chemical Engineering, and a professor of material science & engineering, macromolecular science & engineering and physics.
Anna Simon is supported by the Arnold O. Beckman Postdoctoral Fellowship. This work was also supported by the U.S. Army Research Laboratory, the U.S. Army Research Office, the National Science Foundation and the Welch Foundation.
This is an adaptation of the press release by Marc Airhart of UT Austin.